Construction of the mutant library of OsNramp5
Establishment of error-prone PCR experimental system
Construction of the random mutation library with homologous recombination
Screening of error-prone PCR random mutation library
Determination of Cd2+concentration in solid media
Screening of OsNramp5-mut strains transferring less Cd2+
Screening of OsNramp5-mutstrains transporting Mn2+ normally
Cadmium responding biosensor
Bioinformatics-assisted mutation sites selection
Prediction of OsNramp5 protein topological structure
Prediction of OsNramp5 protein mutation sites
Reference
Establishment of error-prone PCR experimental system.
Recombinant plasmid pFL61-OsNramp5 was constructed by homologous recombination. The mutation frequency of the library was positively correlated with the amount of StarMut Enhancer added into the reaction system. The reaction system contained 0, 1, 2.5, 5, 10, 20 μL StarMut Enhancer to obtain PCR amplification of OsNramp5 cDNA sequence. According to the result of electrophoresis, 10 μL StarMut Enhancer performed the best result. The mutant ratio is about 6.5‰ when 10 μL StarMut Enhancer is added into the reaction system, according to kit instructions. The optimum temperature of error-prone PCR is 63.2 ℃ according to the gradient experiment.
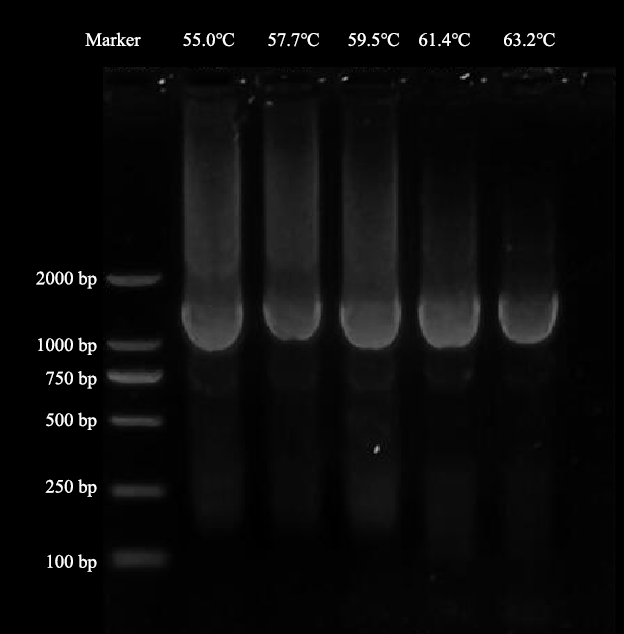
Fig.1 Error-prone PCR results at gradient temperature. Minimal dispersion can be observed at 63.2 °C, while the brightness of the bands indicated a more successful amplification. Therefore, the annealing temperature was set as 63.2 ℃ in the subsequent experiments.
Construction of the random mutation library with homologous recombination.
Linearized pFL61 plasmids and OsNramp5 mutant sequences were used to conduct homologous recombination and built a mutant plasmid library. After the plasmid library was constructed, yeast transformation was performed and screening could be carried out using SD-Ura solid media. Thirteen rounds of plasmids were transformed and about 200 plates were plated in 6 weeks. After 72 hours of incubation in 30 ℃, positive transformants were re-delineated. Twenty-one library plates (Fig.2) were established with a total capacity of about 260 colonies. During our research, we found that the homologous recombinant plasmids showed instability in heredity and low efficiency in the process of transformation. The plasmids also came out with the problem during PCR. We could observe smears or loss of our target bands. We supposed that the simple homologous recombination process lacks the common methylation and phosphorylation which happen normally in common cells, thus decreasing the stability of the plasmids and leading to their breakage during PCR.
Afterwards, to solve this problem, we made attempt to transform the mutant recombinant plasmids into E.coli (Escherichia coli) DH5α for amplifying firstly, and then transferred them into Saccharomyces cerevisiae Δycf1 .The results revealed that the efficiency of transformation was greatly improved. The plates covering 100μL yeast solution showed up to almost 200 CFUs per plate, far exceeding the previous 2 to 30 CFUs. However, after the plasmid amplification in E.coli the number of the same yeast clones turned out to be huge as well. 115 strains were selected randomly to fill in the mutation library.
Fig. 2 Part of the mutant libiaries.
Determination of Cd2+concentration in solid media.
Solid SD-Ura media were set up with different Cd2+ concentration: 0, 30, 40, 50, 100 μM, to find a proper Cd2+concentration. We tried to get a concentration at which the phenotype of yeast transferred to unedited pFL61 plasmid as control group was significantly better than that in HR, the yeast containing OsNramp5-ori. The solid media with too much Cd 2+ led to poor phenotype for pFL61 and HR, while the low Cd2+ concentration in the solid media results in good growth for control and they can't make any difference. In the two occasions mentioned above, we can't decide if the phenotype of yeast in experiment group (OsNramp5-mut yeast) is close to that of Saccharomyces cerevisiae that we inserted in pFL61 or HR (OsNramp5-ori Saccharomyces cerevisiae). Yeasts with a similar phenotype to pFL61 are defined as potential benign mutants while yeasts with phenotypes similar to or worse than HR are considered to contain deleterious mutation sites or null mutation sites. The phenotype difference between pFL61 group and HR group is the most evident when the concentration of Cd2+ is 30 μM. The growth state is suitable for observation after being cultured for 72h. Therefore, the concentration of Cd2+ in the solid SD-Ura media was set as 30 μM in the following experiment.
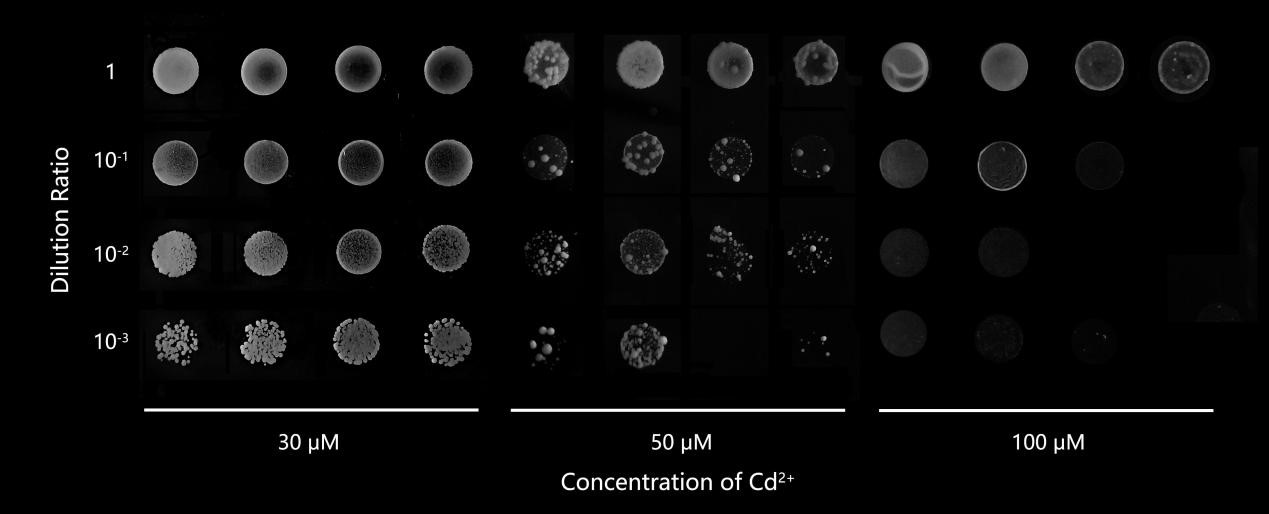
Fig.3 The growth of yeast at different Cd2+ concentrations. The most suitable concentration for screening potential benign mutant strains was 30 μM.
Screening of OsNramp5-mut strains transferring less Cd2+.
Recombinant plasmids were transformed into yeasts, and they could not grow on the solid SD-Ura media without this kind of plasmid. In the process of library construction, we got 260 positive transformants totally. Then, the single colony of each transformant was propagated in liquid SD-Ura media. Different densities of yeast were obtained by gradient dilution, to spot on SD-Ura solid media containing Cd2+. By doing so, the effect of different yeast concentration was further avoided on the final phenotypic observation. 6 mutant strains (Fig.4) were selected by spot experiments, of which the phenotype was close to the control group. Therefore, they were recognized as potential benign mutants. These mutants were further sequenced and analyzed, in order to explore associations among these mutation sites in different mutants. Besides, we also obtained mutants with poorer phenotypes than HR.They may serve as hypersensitive strains of cadmium and may be useful in other future studies on cadmium stress. We concluded that OsNramp5 of these mutants probably increased their ability to transport Cd2+. Therefore, the mutation sites of these mutants were probably related to Cd2+ transportation.
Six mutants with benign phenotype we selected were sequenced, and sequences of two mutants were successfully obtained. Some mutation sites were finally found in OsNramp5 of ep-17: H21L, P22A, Q23P, K26Q, L78Q; in OsNramp5 of ep-11: Q78L, S162G, Q167L, T294A. Due to frame shift mutation, ep5-11 was terminated early at 298th site and ep5-17 was terminated early at the 114 th site. We analyzed that the structure of the protein was damaged resulting in the loss of its ability to transport cadmium, and this change was likely to also affect the transport of Mn 2+.
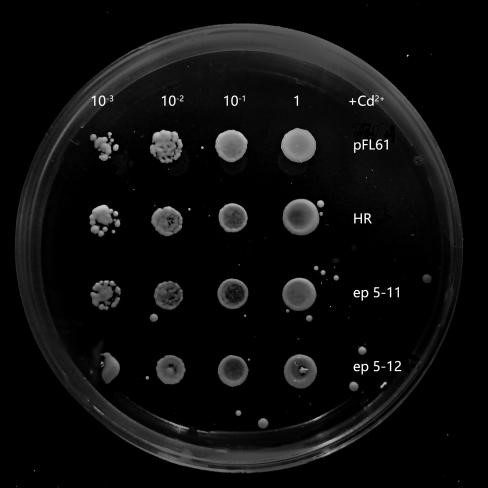
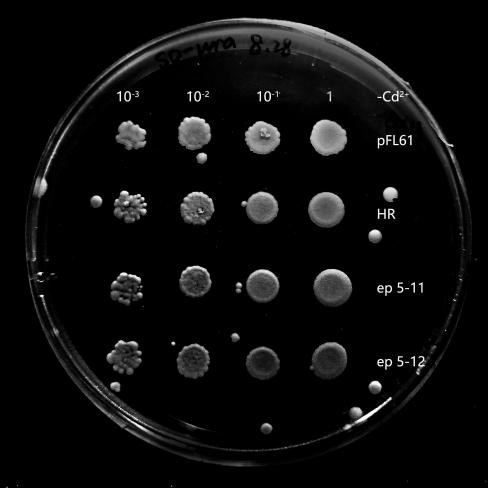
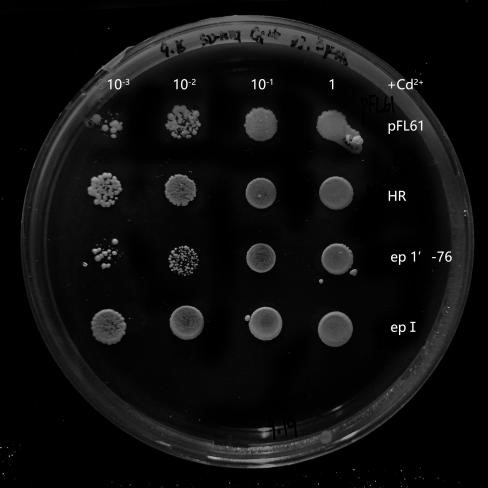
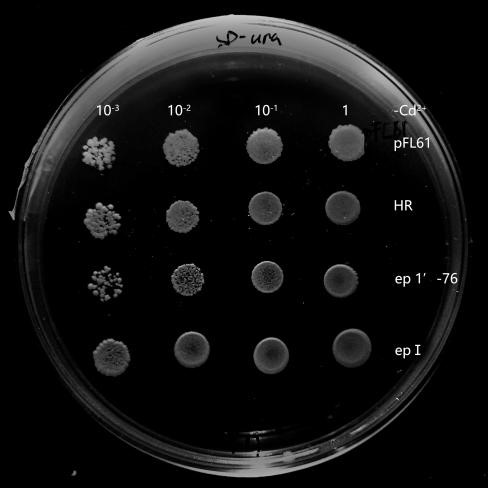
Fig.4 Potential benign mutant strains ep 5-11 and epⅠ. The growth of ep 5-11 and epⅠ on SD-Ura plates contains Cd2+ concentration of 30 μM. It was slightly better than that of pFL61 and HR while they grew similarly to pFL61 and HR on plates without Cd2+, indicating that ep 5-11 and epⅠmay be the potential OsNramp5-mut strains. Its ability of transporting Cd2+ has successfully decreased or their OsNramp5 has been damaged.
Screening of OsNramp5-mut strains transporting Mn2+ normally.
The mutants with reduced transport of Cd2+ selected in primary screening were treated as new experimental materials. They were spotted on Mn2+-deficient SD-Ura media with the control group (Δsmf1-pFL61 and Δsmf1-HR). The Mn2+ in media was specifically chelated by EGTA. As for the mutant strains, Δsmf1-pFL61 could not grow well in the media while Δsmf1-HR can, because the Δsmf1 strain cannot transport Mn 2+ normally, they already had an additional Mn2+ OsNramp5 transporter whose ability of transporting Cd2+ is decreased. If its ability to transport Mn2+ is not affected by the mutation, its growth phenotype should be similar to HR. If the function of OsNramp5 in transporting Mn2+is damaged during the mutation, its growth phenotype should be similar to that of pFL61.
The re-screening strategy using Mn2+ was exactly the same as the primary screening by Cd 2+. Series of densities of yeast solution was set for spot experiment. In conclusion, the strategy to select the benign OsNramp5-mut which can transport Mn2+ normally is that the closer the phenotype of the mutant is to HR, the more likely it is to be the benign mutant strain we expect. Due to the time limitation, it was difficult for us to obtain this part of the data before the paper was submitted, but relevant experiments have been conducted. If the experiments go smoothly, we hope to be allowed to present our final screening results in the thesis defense.
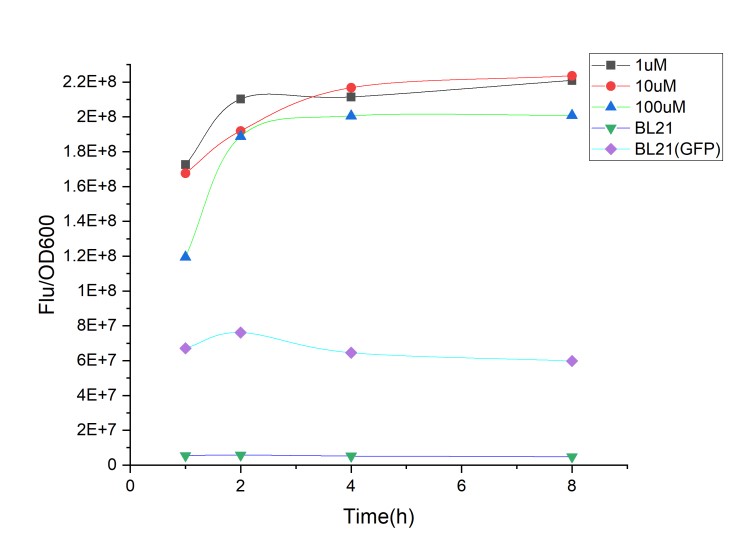
Fig.5 Effect of Cd2+ concentration on engineered bacteria.
Researchers have found that some microorganisms show good ability in Cd2+ resistance and removal (Albano & Macfie, 2016). Especially, Pseudomonas aeruginosa is a heavy-metal resistant bacteria isolated from industry wastewater, which possesses high resistance to Cd 2+. Further researches showed that the Cd2+ resistance ability relies on two metal-recognizing regulatory proteins—P-type ATPase (CadA) and transcription regulatory protein (CadR). The CadR protein has high binding affinity with the promoter of cadR and cadA (P cadR and PcadA) and represses the expression of the CadA genes downstream. However, in the presence of Cd2+, CadR will binding to Cd2+ with high specificity, thus decreases its affinity to its promoter. So this protein acts as a strong repressor of transcription corresponding to the change of Cd2+ concentration.
In our project, we constructed a gene circuit with cadR, PcadA and green fluorescent protein (GFP) in Escherichia coli. The GFP fluorescence intensity may change according to the Cd2+ concentration, the higher the Cd2+ concentration is, the stronger the fluorescence intensity may be, hoping that it can act as a microbial biosensor to present the intracellular cadmium concentration which affected by different OsNramp5.
The characterization experiment showed that, the E.coil BL21 which contained recombinant plasmids got higher fluorescence intensity compared with non-loaded BL21 and BL21 which transformed plasmids with GFP only. At around 2 hours, the fluorescence intensity of engineered bacteria in 10 μM Cd2+ solution was higher than that in 1 μM Cd2+ solution, since the bacteria needed some time to adapt to the environment of Cd2+. But in general, there was a positive correlation between Cd2+ concentration and GFP fluorescence intensity. However, the engineered bacteria was less responsive to 100 μM Cd2+ comparatively. It was probably because the engineered bacteria exhibited lower resistance to cadmium when the concentration is too high, since the physicochemical properties of the bacteria changed or part of the GFP protein denatured.
Directed evolution, by establishing mutation libraries and using high-throughput screening methods, is a common strategy for protein design and modification in protein engineering, which can rapidly improve the specific properties of the protein. Directed evolution brings infinite possibilities for protein modification. Discontinuous directed evolution relying on traditional methods often results in huge mutation libraries, consuming a lot of time and energy, ending up with bad results obtained often [1]. In recent years, with the improvement of computer power and the optimization of algorithms, the computer-assisted directed evolution for protein has developed quickly, which has opened up a new direction for directed evolution. Protein computational design based on structural simulation and energy calculation assisting to select mutation site provides insights into "precise" directed evolution[2]. Therefore, bioinformatics analysis can help to reduce the redundant work for the construction of discontinuous directed evolutionary mutation scheme. In this study, the error-prone PCR strategy was adopted to directionally evolve OsNramp5. Due to its large error-prone mutation library and unconcentrated mutation sites, the ideal result was not obtained. We intend to use this method to analyze our target protein and provide theoretical reference for other researchers.
Prediction of OsNramp5 protein topological structure
OsNramp5 is a typical membrane protein[3]. The topological structure of a membrane protein can be defined as its transmembrane way, the position of the membrane segment in the amino acid sequence and the overall orientation of the protein molecule on the membrane. The secondary structure of membrane protein can be represented by its topological structure, which can be regarded as the intermediate of membrane protein folding, and is the starting point for predicting the tertiary structure of membrane protein. In order to learn more about OsNramp5 protein structure, its protein topology structure is predicted through TMHMM v2.0 (http//www.cbs.dtu.dk/services/TMHMM/). And it turns out that OsNramp5 has 12 typical transmembrane helical regions and 76.3% of the protein terminal are in the membrane, which is structurally similar to other Nramp families [4](Table S2).
Prediction of OsNramp5 protein mutation sites
Mutation hotspots of the OsNramp5 have been analyzed by searching related papers and comparing with structure of channel proteins from the same family that have been analyzed. The sites 60D, 63N, 157G, 180E and 235M were thought to be important for absorbing Mn2+ in OsNramp5. Different mutations of 235M result in different states of absorbing Mn2+ and Cd2+[5,6]. The sites 60D and 63N are located outside membrane domain; the sites 157G, 180E and 235M are respectively located in S4, S5 and S6 transmembrane domain. "DPGN" located in the first transmembrane domain is a conserved domain (Table S3), and is thought to be an important domain interacting with Mn2+ homologous modeling for OsNramp5 by SWISS-MODEL[5,7-10]. The model is based on the transmembrane protein of Eremococcus coleocola MntH (PDB Code: 6TL2.1). This figure is made by PyMOL (Fig.7). These potential mutation sites are all from the previous reports or software prediction. Accuracy and specific effects of these sites in OsNramp5 have not been tested, and only taken as reference.
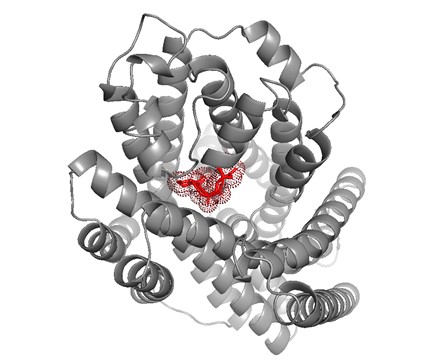
Fig.7 OsNramp5 metal transporter.The highly conserved "DPNG" region, located at the c-terminus of the first transmembrane region, is consistent with the calibration of 12 cadmium transmembrane residues in Nramp family homologue by Gros et al. OsNramp5 homologous model was based on Eremococcus coleocola MntH (PDB Code: 6TL2.1) transmembrane protein, and its potential metal ion action region was labeled, which was used as the preferred mutation site.
[1] SHELDON R A, PEREIRA P C. Biocatalysis engineering: the big picture [J]. Chemical Society Reviews, 2017, 46(10): 2678-91.
[2] YANG K K, WU Z, ARNOLD F H. Machine-learning-guided directed evolution for protein engineering [J]. Nature Methods, 2019, 16(8): 687-94.
[3] MANI A, SANKARANARAYANAN K. In Silico Analysis of Natural Resistance-Associated Macrophage Protein (NRAMP) Family of Transporters in Rice [J]. The Protein Journal, 2018, 37(3): 237-47.
[4] BOZZI A T, GAUDET R. Molecular Mechanism of Nramp-Family Transition Metal Transport [J]. J Mol Biol, 2021, 433(16): 31.
[5] POTTIER M, OOMEN R, PICCO C, et al. Identification of mutations allowing Natural Resistance Associated Macrophage Proteins (NRAMP) to discriminate against cadmium [J]. Plant Journal, 2015, 83(4): 625-37.
[6] JIYU L. Structure and function of Arabidopsis thaliana metal transporter NRAMPs [D]; Nanjing Agricultural University, 2018.
[7] BIENERT S, WATERHOUSE A, DE BEER TJAART A P, et al. The SWISS-MODEL Repository—new features and functionality [J]. Nucleic Acids Research, 2016, 45(D1): D313-D9.
[8] BERTONI M, KIEFER F, BIASINI M, et al. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology [J]. Scientific Reports, 2017, 7(1): 10480.
[9] STUDER G, REMPFER C, WATERHOUSE A M, et al. QMEANDisCo—distance constraints applied on model quality estimation [J]. Bioinformatics, 2019, 36(6): 1765-71.
[10] STUDER G, TAURIELLO G, BIENERT S, et al. ProMod3—A versatile homology modelling toolbox [J]. PLOS Computational Biology, 2021, 17(1): e1008667.
Download:
Supplementary Information-1